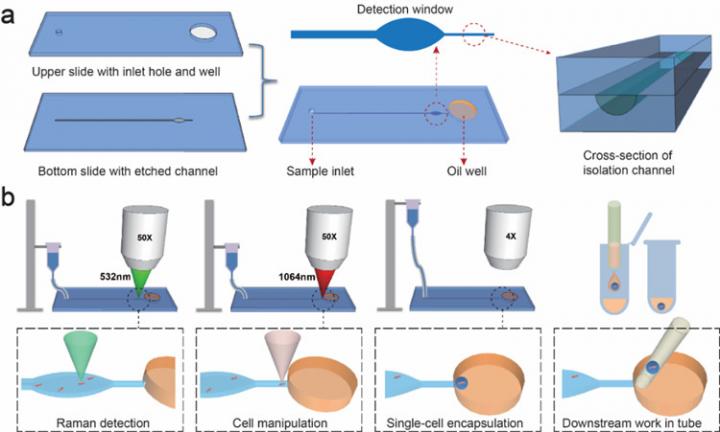
IMAGE: Design of the RAGE chip
view more
Credit: XU Teng
A new function-based sequencing technique using optical tweezers and taking advantage of the properties of gravity is letting researchers analyze bacteria cells one by one. The study, conducted by researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences, was published in Small on June 9.
Bacteria cells are so teeny tiny that it has been very difficult to analyze just one bacterial cell, or bacterium, at a time. As a result, lots of them, sometimes millions at a time, have to be analyzed simultaneously. This tells us a lot about the group as a whole, but it prevents researchers from being able to investigate the link between a single bacterium’s genotype, or complete set of genes, and its phenotype, or the set of characteristics that result from the interaction of its genes and the environment.
A simple way to think about the distinction between genotype and phenotype is to note that while a single corn plant’s genotype might allow it to grow three feet tall, if not much fertilizer is applied, then the corn plant’s phenotype might be that it only grew to be two feet tall.
Analysis of the link between genotype and phenotype is straightforward for such a large organism, and very useful too.
Similar insights about the genotype-phenotype relationship of a single bacterium, not least with respect to infectious disease, have long been sought but hindered by a bacterium’s size, which are typically just a few millionths of a meter in length.
Researchers from the Single-Cell Center at QIBEBT have developed a bacteria-profiling technique called Raman-Activated Gravity-driven single-cell Encapsulation and Sequencing, or RAGE sequencing. In the technique, the phenotypes of individual cells are analyzed one by one, then carefully packaged in a ‘picoliter microdroplet’ (a trillionth of a liter) that is exported and indexed in a one cell per test-tube manner ready for gene sequencing later on.
The process involves a RAGE ‘chip’ of two quartz layers bonded together and that have an inlet hole, oil well, and micro-channel etched into them. ‘Optical tweezers’, or a highly focused laser beam that produces an attractive or repulsive force, manipulates the bacterium in liquid through the channel, assisted by gravity.
The form, structure and metabolic features of the bacterium – essentially its phenotype – are then investigated via a detection window using ‘Raman spectroscopy’, an analytical technique that exploits the interaction of light with the chemical bonds within a material.
“Finally the bacterium is encapsulated in the microdroplet, which is then transferred to a tube for gene sequencing or cultivation of the cell,” said Prof. MA Bo, corresponding author of the study.
The microdroplet packaging is extremely important, as it allows the very small amount of DNA in a single bacterial cell to be amplified in a very even way, a key challenge for decoding its genome fully, according to XU Teng, a graduate student on the team that developed the method.
“We are able to, directly from a urine sample, obtain antibiotic resistance features and an essentially complete genome sequence simultaneously from precisely one cell. This offers the highest possible resolution for bacterial diagnosis and drug treatment,” said Prof. XU Jian, another corresponding author of the study.
Based on this technology, the researchers have developed an instrument called CAST-R to support rapid antibiotic selection and genome sequencing of pathogens, all at the level of one cell. This instrument means much faster and more precise antibiotic treatment, and much higher sensitivity in tracking and fighting bacterial antibiotic resistance, which is a major threat to the future of human society.
###
TDnews














