[ad_1]
The complex 3D structure of one of the world’s most lethal families of plant viruses has been revealed in unprecedented detail by scientists at the UK’s University of Leeds.
Geminiviruses are responsible for diseases affecting crops such as cassava and maize in Africa, cotton in the Indian subcontinent and tomatoes across Europe.
Being able to see its structrure in great detail is vital as it could help virologists and molecular biologists better understand its lifecyle, and develop new ways to stop the spread of these viruses and the diseases they cause.
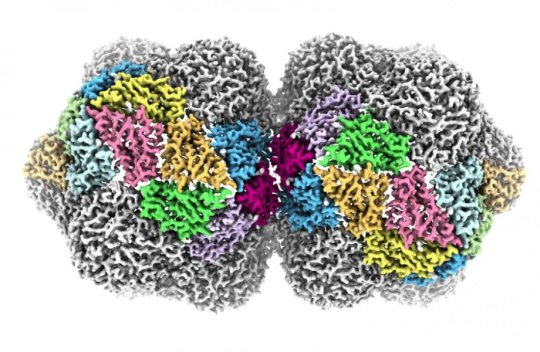
The viruses are named for their curious shape. Viruses usually have a protective shell of protein, or a capsid, that acts to protect their genetic material in the environment. In most viruses, this capsid is roughly spherical, but the geminivirus has a ‘twinned’ capsid formed by two roughly spherical shapes fused together.
The molecular details of how this twinned capsid is achieved — and how it assembles in cells or expands to release the genome and start a new infection — has remained a mystery, despite the risk posed by the virus to agricultural economies worldwide.
Researchers at the University’s Astbury Centre for Structural Molecular Biology used cryo electron microscopy techniques to study geminivirus structure at undprecedented resolution, and in the process have begun to untangle its assembly mechanisms.
Published in Nature Communications, the study reveals how the capsid of the geminivirus is built and how its single-stranded DNA genome is packaged.
“In many other types of virus, the spherical capsids are built from a single protein that adopts three different shapes, which then fit together to form a closed container,” explains Professor Neil Ranson, who led the research team at the Astbury Centre. “But geminivurses are not spherical, so must be using a different set of rules. Using cryo-EM, we’ve been able to show that they do use three different shapes of the same protein, but with a completely different rulebook for assembly.”
One of the difficulties in studying geminviruses is growing them in sufficient quantities for structural studies. The team studied a type of geminvirus called ageratum yellow vein virus, which was produced in tobacco plants under carefully controlled conditions by researchers at the John Innes Centre in Norwich.
The team at the John Innes Centre, led by Dr Keith Saunders and Professor George Lomonossoff, also developed a method for assembling geminivirus particles within plants in the absence of infection. This highlighted the role played by the single-stranded DNA in particle formation.
“Having worked for many years to understand the diseases geminiviruses cause, it was very satisfying to apply modern genetic methods to generate these geminate structures,” said Dr Saunders.
“We’ve now been able to analyse the role that different conformations of the coat protein play in particle assembly, and we can potentially make other viruses and virus-like particles that might otherwise be impossible to isolate from natural infections.”
“Using our ‘next generation’ cryo electron microscopy we have modelled the position of the majority of the atoms in the virus,” said Dr Emma Hesketh, a post-doctoral researcher in the Astbury Centre, who carried out the work to create the images of the structure.
“This technology is often referred to as the resolution revolution, and it’s enabled us to get this fascinating — and very beautiful — insight into these structures. By using these techniques to understand the structure and the life cycle of these viruses, we can come a step closer to understanding how to interrupt that life cycle, and inhibit the spread of plant disease.”
Story Source:
Materials provided by University of Leeds. Note: Content may be edited for style and length.
[ad_2]