[ad_1]
A comprehensive analysis of sewage collected in 74 cities in 60 countries has yielded the first, comparable global data, which show the levels and types of antimicrobial resistant bacteria that are present in mainly healthy people in these countries. The National Food Institute, Technical University of Denmark, headed the study, which was conducted by an international team of researchers.
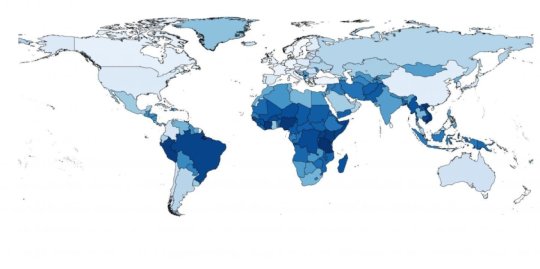
In a metagenomics study, the researchers have mapped out all the DNA material in the sewage samples and found that according to antimicrobial resistance the world’s countries fall within two groups. North America, Western Europe, Australia and New Zealand generally have the lowest levels of antimicrobial resistance, while Asia, Africa and South America have the highest levels.
Brazil, India and Vietnam have the greatest diversity in resistance genes, while Australia and New Zealand have the lowest.
Sanitation and health closely linked to antimicrobial resistance
According to the researchers, the use of antimicrobials only explains a minor part of the occurrence of antimicrobial resistance in the various countries. Therefore, the researchers have searched for other factors that could be either drivers for or indicators of the occurrence of resistant bacteria. In this work, they have used several comprehensive data sets from the World Bank, which e.g. have measured the countries’ health status and stage of development.
The researchers’ work shows that most of the variables, which are associated with the occurrence of antimicrobial resistance in a country, are related to the sanitary conditions in the country and the population’s general state of health.
“In the fight against antimicrobial resistance, our findings suggest that it would be a very effective strategy if concerted efforts were made to improve sanitary conditions in countries with high levels of antimicrobial resistance,” Professor Frank Aarestrup from the National Food Institute and head of the study says.
Using the same data from the World Bank, the researchers have also predicted the levels of antimicrobial resistance in 259 countries/territories and they have drawn up a world map of resistance in healthy populations. According to their estimates, the Netherlands, New Zealand and Sweden have the lowest levels of resistance, whereas Tanzania, Vietnam and Nigeria have the highest levels. According to the estimates, Denmark has the sixth lowest level of antimicrobial resistance.
Reusable data
Contrary to data from traditional methods of analysis, raw data from metagenomics studies can be reused to examine other problems. The researchers from the sewage project are e.g. using data from the study to analyze the occurrence of other disease-causing microorganisms in the sewage.
As more resistance genes emerge in the future, researchers will also be able to reanalyse publicly available raw data from metagenomics studies to quickly uncover how these genes have emerged and spread.
A step closer to a global surveillance system
The researchers will use the experience gained from the project to fulfill their overall ambition of developing a worldwide surveillance system that can continuously monitor the occurrence and spread of disease-causing microorganisms and antimicrobial resistance.
“Analysing sewage can quickly and relatively cheaply show exactly which bacteria abound in an area — and collecting and analysing sewage doesn’t require ethical approval, as the material cannot be traced back to individuals. Both parameters help to make a surveillance system via sewage a viable option — also in developing countries,” Professor Frank Aarestrup explains.
The researchers’ ambition is to develop a system, which makes it possible to exchange and interpret information in real time. As such, it would be possible to use the global surveillance data e.g. to manage diseases that threaten to spread beyond a country’s borders and develop into pandemics, such as Ebola, measles, polio or cholera.
Story Source:
Materials provided by Technical University of Denmark. Original written by Miriam Meister. Note: Content may be edited for style and length.
[ad_2]