[ad_1]
The nuclei of cells are never static, even when the chromosomes they contain appear to be at rest. Theorists at Rice University have detailed the combination of forces that drive their constant motion.
An extension of their landmark energy landscape model for proteins helped researchers at Rice’s Center for Theoretical Biological Physics form a theory that unifies the structural and dynamic aspects of the genome during interphase, the time before reproduction when nuclear DNA conducts the symphony of activity that keeps the cell humming.
Scientists have long been able to watch cells go through their cycle from interphase to mitosis and back, but it has been difficult to understand what’s happening during each phase. In the new study, the Rice researchers determined that the energy landscape model they developed to analyze the spatial folding of chromosomes could also account for genome dynamics that experimentalists have recently observed during interphase.
The open access paper published today in the Proceedings of the National Academy of Sciences by physicists José Onuchic and Peter Wolynes and postdoctoral researchers Michele Di Pierro and Davit Potoyan draws upon the lab’s long experience in modeling how proteins fold.
Those models rely on coarse-grained algorithms in which a subset of amino acids in a chain — either a protein or DNA — represents the entire chain. The inherent energies of the individual acids determine how the chain will fold and how likely they are to associate with other beads along the chain.
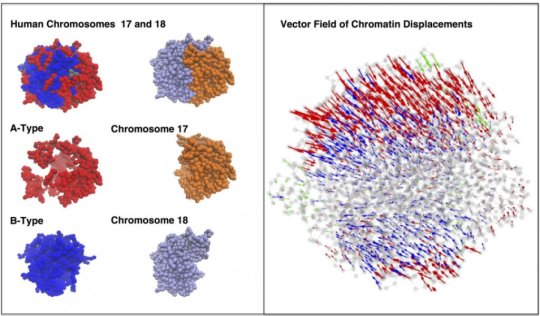
Over the past four years, the researchers have refined their algorithms into their Minimal Chromatin Model, which simulates the energy landscape of DNA as it divides, condenses into a tight blob during interphase and then blossoms into the familiar X- and Y-shaped chromosomes in preparation for mitosis.
The model shows that both random diffusion driven by Brownian motion and more directed movements triggered by molecular motors help DNA wriggle in its nuclear sphere as it continually reorganizes itself.
“At the molecular scale, the ordinary thermal motion of particles is enough to move them around and get them out of jams where they’re too strongly bound to other particles,” Wolynes said. “Typically, they go to the state that’s the most thermodynamically stable. For protein folding, these thermal motions are paramount, but in the macroscopic world, they are completely inadequate.
“The problem at the scale of the chromosome is that it’s much smaller than macroscopic things we see around us but much larger than a molecule. So which rules are we supposed to apply?”
The answer turned out to be both.
Wolynes noted that experiments had showed diffusion-like motion within the chromosome, but also evidence that ATP-driven molecular motors were helping direct things. Experiments by other labs that stopped the motors also stopped some of the changes in chromosomal structure.
“We decided to see what happens if our model assumes the movements are equivalent to those at equilibrium but are being stirred about by motors that more or less do what the random thermal motions do, but more violently,” he said. “That got us to something that was very much in agreement with experiments.”
The simulated interactions between two chromosomes in a lymphoblastoid cell also revealed the presence of between 10 and 20 dynamically associated domains (DADs), blobs of compartmentalized chromatin — segments of DNA and their associated epigenetic markers and other proteins — that tended to move in a coherent way. The researchers argue that DADs arise from the microscale phase separation, like oil and water, of chromatin segments that have different biochemical properties.
“These DADs are a consequence of a competition between the tendency to phase-separate and the constraints that arise from the polymer nature of chromatin,” Onuchic said. Unlike oil and water that fully separate, chromatin creates multiple domains that segregate by type, he said.
“We found locations in the chromosome that deviate from the diffusion law, as some of them diffuse faster than others,” Wolynes said. “These jelly droplets are all different sizes. Some diffuse for a long time, some for a shorter time, and there’s a threshold that defines these modes of motion.”
He compared them to mayonnaise, in which the proteins in egg yolk form droplets that bind oil and water. “Phase separation in chromatin makes the chromosome into a kind of salad dressing in which droplets with different epigenetic marks move coherently,” Wolynes said.
Di Pierro said the work should appeal to both biologists who study the structure of DNA and the biophysicists who study its movement. “We want our theory to bridge that gap between dynamics and structure,” he said. “With a bit of physics and a bit of engineering, we can actually show that what looks weird from one point of view or the other all fits into the same picture.”
[ad_2]