[ad_1]
Allmost half of our DNA sequences are made up of jumping genes — also known as transposons. They jump around the genome in developing sperm and egg cells and are important to evolution. But their mobilization can also cause new mutations that lead to diseases, such as hemophilia and cancer. Remarkably little is known about when and where their movements occur in developing reproductive cells, the key process that ensures their propagation in future generations, but can lead to genetic disorders for the hosts.
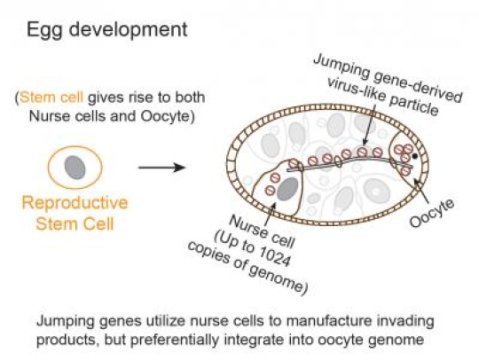
To address this problem, a team* of Carnegie researchers developed new techniques to track the mobilization of jumping genes. They found that during a particular period of egg development, a group of jumping-genes called retrotransposons hijacks special cells called nurse cells that nurture the developing eggs. These jumping genes use nurse cells to produce invasive material (copies of themselves called virus-like particles) that move into a nearby egg and then mobilize into the egg’s DNA. The research is published in the July 26 on-line issue of Cell.
Animals have unwittingly developed a powerful system to suppress jumping gene activity that uses small, non-coding RNAs called piRNAs, which recognize jumping genes and suppress their activity. Occasionally, jumping genes still manage to move, suggesting that they employ some special tactics to escape piRNA control. However, tracking the mobilization of jumping genes to understand their tactics has been a daunting task.
The Carnegie team developed approaches to track the movements of jumping genes using the fruit fly Drosophila melanogaster. To facilitate their investigation, they disrupted piRNA suppression to increase the activity of these jumping genes and then monitored the movement of them during the egg-development process. This led to their discovery on the tactic that allows jumping genes to move.
Carnegie co-author Zhao Zhang explained: “We were very surprised that the these jumping genes barely moved in stem cells that produce developing egg cells, possibly because the stem cells would only have two copies of the genome for these jumping genes to use. Instead, these moving elements used the supporting nurse cells, which could provide up to thousands copies of the genome per cell, as factories to massively manufacture virus-like particles capable of integration. However, they didn’t integrate into nurse cells where they were produced. Rather, they waited while they were transported into an interconnected egg cell, and then added hundreds, if not thousands, of new copies of themselves into the egg DNA. Our research shows how parasitic genetic elements can time their activity and distinguish between different cell types to robustly propagate to drive evolutionary change and cause disease.”
“My group has found that egg development in mammals uses many of the same mechanisms as in the fruit fly, such as feeding the developing egg using nurse cells. So the Zhang group’s findings are likely to be important for understanding mammalian evolution and disease as well,” commented Allan Spradling, who is a pioneer researcher on studying the egg development in both fruit fly and mammals and a longtime scientist at Carnegie’s Department of Embryology.
“Since Carnegie’s Barbara McClintock made the seminal contribution of discovering the jumping genes more than six decades ago, we have not been able to understand how they mobilize as animals prepare for their next generation. Now the junior research group lead by our Staff Associate, Zhao Zhang, has made an important step toward solving this long-standing puzzle,” remarked Yixian Zheng, the director of Carnegie’s Department of Embryology.
Story Source:
Materials provided by Carnegie Institution for Science. Note: Content may be edited for style and length.
[ad_2]