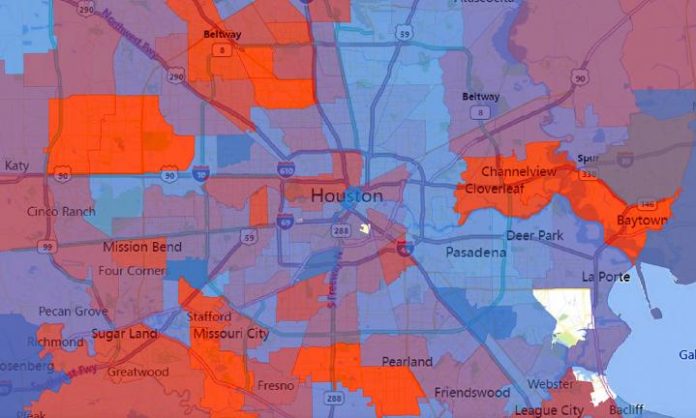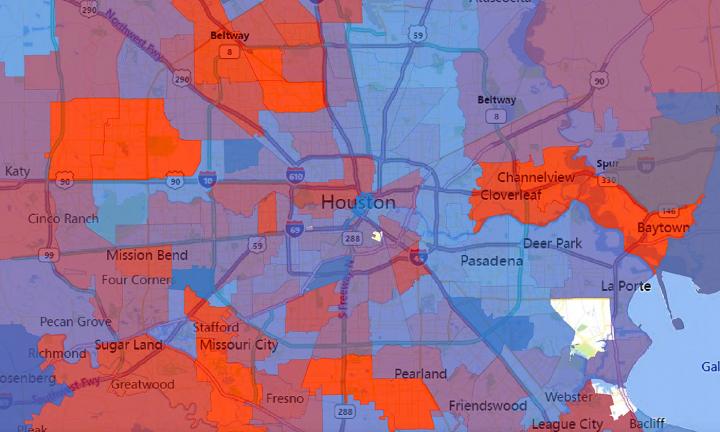
Photo: The number of virus strains present in each zip code in Houston during the second wave of COVID-19 cases in summer 2020. Number of strains is represented by a spectrum…
view more
Credit Image: Houston Methodist/University of Texas at Austin
A study involving more than 5,000 COVID-19 patients in Houston finds that the virus that causes the disease is accumulating genetic mutations, one of which may have made it more contagious. According to the paper published in the peer-reviewed journal mBIO, that mutation, called D614G, is located in the spike protein that pries open our cells for viral entry. It’s the largest peer-reviewed study of SARS-CoV-2 genome sequences in one metropolitan region of the U.S. to date.
The paper shows “the virus is mutating due to a combination of neutral drift — which just means random genetic changes that don’t help or hurt the virus — and pressure from our immune systems,” said Ilya Finkelstein, associate professor of molecular biosciences at The University of Texas at Austin and co-author of the study. The study was carried out by scientists at Houston Methodist Hospital, UT Austin and elsewhere.
During the initial wave of the pandemic, 71% of the novel coronaviruses identified in patients in Houston had this mutation. When the second wave of the outbreak hit Houston during the summer, this variant had leaped to 99.9% prevalence. This mirrors a trend observed around the world. A study published in July based on more than 28,000 genome sequences found that variants carrying the D614G mutation became the globally dominant form of SARS-CoV-2 in about a month. SARS-CoV-2 is the coronavirus that causes COVID-19.
So why did strains containing this mutation outcompete those that didn’t have it?
Perhaps they’re more contagious. A study of more than 25,000 genome sequences in the U.K. found that viruses with the mutation tended to transmit slightly faster than those without it and caused larger clusters of infections. Natural selection would favor strains of the virus that transmit more easily. But not all scientists are convinced. Some have suggested another explanation, called “founder’s effects.” In that scenario, the D614G mutation might have been more common in the first viruses to arrive in Europe and North America, essentially giving them a head start on other strains.
The spike protein is also continuing to accumulate additional mutations of unknown significance. The Houston Methodist-UT Austin team also showed in lab experiments that at least one such mutation allows spike to evade a neutralizing antibody that humans naturally produce to fight SARS-CoV-2 infections. This may allow that variant of the virus to more easily slip past our immune systems. Although it is not clear yet whether that translates into it also being more easily transmitted between individuals.
The good news is that this mutation is rare and does not appear to make the disease more severe for infected patients. According to Finkelstein, the group did not see viruses that have learned to evade first-generation vaccines and therapeutic antibody formulations.
“The virus continues to mutate as it rips through the world,” Finkelstein said. “Real-time surveillance efforts like our study will ensure that global vaccines and therapeutics are always one step ahead.”
The scientists noted a total of 285 mutations across thousands of infections, although most don’t appear to have a significant effect on how severe the disease is. Ongoing studies are continuing to surveil the third wave of COVID-19 patients and to characterize how the virus is adapting to neutralizing antibodies that are produced by our immune systems. Each new infection is a roll of the dice, an additional chance to develop more dangerous mutations.
“We have given this virus a lot of chances,” lead author James Musser of Houston Methodist told The Washington Post. “There is a huge population size out there right now.”
Several other UT Austin authors contributed to the work: visiting scholar Jimmy Gollihar, associate professor of molecular biosciences Jason S. McLellan and graduate students Chia-Wei Chou, Kamyab Javanmardi and Hung-Che Kuo.
The UT Austin team tested different genetic variants of the virus’s spike protein, the part that allows it to infect host cells, to measure the protein’s stability and to see how well it binds to a receptor on host cells and to neutralizing antibodies. Earlier in the year, McLellan and his team at UT Austin, in collaboration with researchers at the National Institutes of Health, developed the first 3D map of the coronavirus spike protein for an innovation that now factors into several leading vaccine candidates’ designs.
The researchers found that SARS-CoV-2 was introduced to the Houston area many times, independently, from diverse geographic regions, with virus strains from Europe, Asia, South America and elsewhere in the United States. There was widespread community dissemination soon after COVID-19 cases were reported in Houston.
This study was supported by the Fondren Foundation, Houston Methodist Hospital and Research Institute, the National Institutes of Health, the National Institute of Allergy and Infectious Diseases, the Welch Foundation, the National Science Foundation and the Defense Advanced Research Projects Agency. Ilya Finkelstein is a CPRIT scholar in cancer research, funded by the Cancer Prevention and Research Institute of Texas.
An earlier version of the paper was posted last month to the preprint server medRxiv.
###
TDnews (tunisiesoir.com)