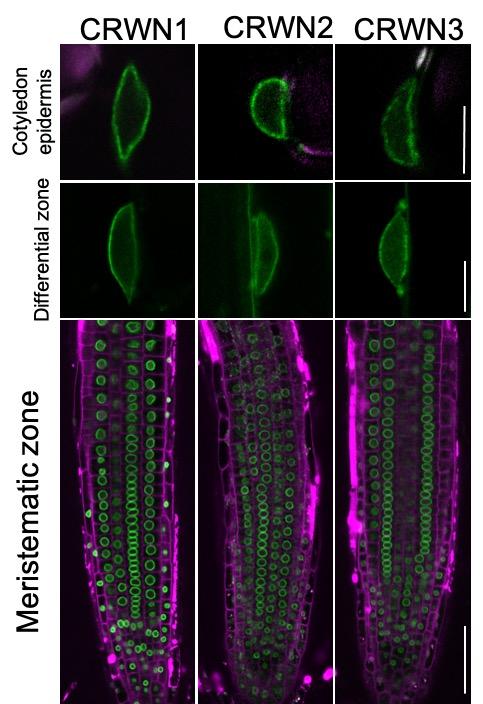
Photo: Researchers at the University of Tokyo have confirmed that crowded nuclei proteins (CRWN1-3) support the oval shape of plant cell nuclei and also have a role in regulating the expression…
view more
Credit Image: Image by Yuki Sakamoto, CC BY, first published in Nature Communications.
Researchers at the University of Tokyo have identified how the architecture of the cell nucleus can change gene activity in plants. This discovery reveals fundamental knowledge about genome regulation and points towards future methods for potentially manipulating the expression of many genes simultaneously.
The long strands of DNA and the protein machinery needed to turn gene expression on or off are contained, floating within the nuclei of cells. The nucleus is essentially a sack made of a flexible, double-membrane envelope that is supported by an inner, fine-mesh frame of proteins called the nuclear lamina.
“DNA does not drift aimlessly within the nucleus. We expect that there is nonrandom spatial positioning of genes around the nuclear lamina,” said Professor Sachihiro Matsunaga who led the research project from the University of Tokyo Graduate School of Frontier Sciences, recently published in Nature Communications.
Gene regulation is often studied at the one-dimensional level of reading the DNA sequence. Additional layers of gene regulation exist in 3D by changing the shape of the DNA strand. Examples include the epigenetic code that dictates how tightly to wind up the strands of DNA and the phenomenon of “kissing genes,” where distant segments of the DNA strand fold together and change the activity of the genes that touch each other.
These new results provide evidence for another 3D method of gene regulation involving not just the architecture of the genome, but the architecture of its container, the nucleus.
The scientific community has long known that the shape and size of the nucleus can fluctuate dramatically during a cell’s life and that these changes can even be timed as an “internal clock” to determine the age of a cell. However, these discoveries have been made using animal cells. Plants do not possess any genes evolutionarily related to the genes responsible for the nuclear lamina in animals.
“Textbooks usually have a few sentences about animal lamina, but nothing at all to say about plant lamina,” said Matsunaga.
Prior work in 2013 by some members of the research team identified a group of four proteins known as CROWDED NUCLEI (CRWN) as the most likely components of the plant nuclear lamina.
To confirm the presence of CRWN proteins in the lamina, researchers first attached fluorescent tags onto the proteins and isolated nuclei out of root cells from young thale cress plants, the roadside weed commonly used in research labs. Then they measured the proteins’ location in ultrahigh-resolution microscopy images.
These extremely zoomed-in images show weblike patterns formed by the CRWN proteins around the shell of the nucleus.
Healthy plant cells have an oval-shaped nucleus, looking like a large egg in the center of the cell. Plants genetically altered to lack CRWN proteins have nuclei that are smaller and rounder than normal, likely creating a more crowded environment for the DNA inside.
Researchers then screened the genetically altered plants to see if any other genes had different activity levels when crwn genes were inhibited. Multiple genes known to be involved in responding to copper were less active, indicating that somehow the nuclear lamina is connected to copper tolerance.
Plants that lack CRWN proteins grow shorter than healthy plants even in normal soil. Thale cress with inactive crwn genes planted in soil with high copper levels grew even smaller with a significantly weaker appearance, further evidence that the nuclear lamina has a role in plants’ response to environmental stress.
Researchers also visualized the physical location of copper tolerance genes within the nucleus of both normal and high copper levels. In healthy plants in the high copper condition, the copper tolerance genes clustered together and moved even closer to the periphery of the nucleus. The copper tolerance genes appeared to spread out and drift around the nuclei in plants with inactive crwn genes.
“If the plant nucleus has distinct regions for active transcription of DNA, it is likely that those regions will be near the nuclear lamina. This is important and interesting because it is opposite to animal cells, which we know have active regions in the center of nuclei while the periphery is inactive,” said Matsunaga.
Most gene editing technologies to increase or decrease gene activity work directly at the one-dimensional level of altering the DNA sequence of the individual gene. Understanding how the nuclear lamina affects gene expression could reveal future methods for altering the activity of many genes at the same time by resculpting the genome and nuclear lamina.
###
Research Article
Yuki Sakamoto, Mayuko Sato, Yoshikatsu Sato, Akihito Harada, Takamasa Suzuki,
Chieko Goto, Kentaro Tamura, Kiminori Toyooka, Hiroshi Kimura, Yasuyuki
Ohkawa, Ikuko Hara-Nishimura, Shingo Takagi, Sachihiro Matsunaga. 24 Nov 2020. Subnuclear gene positioning through lamina association affects copper tolerance. Nature Communications. DOI: 10.1038/s41467-020-19621-z
https:/
Related Links
Matsunaga Lab: http://park.
Graduate School of Frontier Sciences: https:/
Researcher Contacts
Professor Sachihiro Matsunaga
Department of Integrated Biosciences, Graduate School of Frontier Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033 Japan
Tel: +81-04-7136-3706
Email: sachi@edu.k.u-tokyo.ac.jp
Press officer contact
Ms. Caitlin Devor
Division for Strategic Public Relations, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 133-8654, JAPAN
Tel: +81-080-9707-8178
E-mail: press-releases.adm@gs.mail.u-tokyo.ac.jp
About the University of Tokyo
The University of Tokyo is Japan’s leading university and one of the world’s top research universities. The vast research output of some 6,000 researchers is published in the world’s top journals across the arts and sciences. Our vibrant student body of around 15,000 undergraduate and 15,000 graduate students includes over 4,000 international students. Find out more at http://www.
TDnews (tunisiesoir.com)














