Scientists have discovered hundreds of unusually large, bacteria-killing viruses with capabilities normally associated with living organisms, blurring the line between living microbes and viral machines.
These phages — short for bacteriophages, so-called because they “eat” bacteria — are of a size and complexity considered typical of life, carry numerous genes normally found in bacteria and use these genes against their bacterial hosts.
University of California, Berkeley, researchers and their collaborators found these huge phages by scouring a large database of DNA that they generated from nearly 30 different Earth environments, ranging from the guts of premature infants and pregnant women to a Tibetan hot spring, a South African bioreactor, hospital rooms, oceans, lakes and deep underground.
Altogether they identified 351 different huge phages, all with genomes four or more times larger than the average genomes of viruses that prey on single-celled bacteria.
Among these is the largest bacteriophage discovered to date: Its genome, 735,000 base-pairs long, is nearly 15 times larger than the average phage. This largest known phage genome is much larger than the genomes of many bacteria.
“We are exploring Earth’s microbiomes, and sometimes unexpected things turn up. These viruses of bacteria are a part of biology, of replicating entities, that we know very little about,” said Jill Banfield, a UC Berkeley professor of earth and planetary science and of environmental science, policy and management, and senior author of a paper about the findings appearing Feb 12 in the journal Nature. “These huge phages bridge the gap between non-living bacteriophages, on the one hand, and bacteria and Archaea. There definitely seem to be successful strategies of existence that are hybrids between what we think of as traditional viruses and traditional living organisms.”
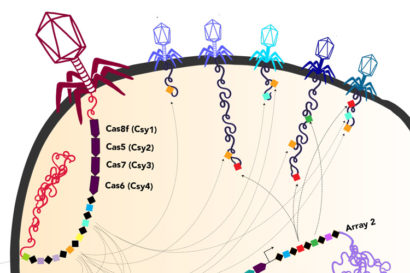
Ironically, within the DNA that these huge phages lug around are parts of the CRISPR system that bacteria use to fight viruses. It’s likely that once these phages inject their DNA into bacteria, the viral CRISPR system augments the CRISPR system of the host bacteria, probably mostly to target other viruses.
“It is fascinating how these phages have repurposed this system we thought of as bacterial or archaeal to use for their own benefit against their competition, to fuel warfare between these viruses,” said UC Berkeley graduate student Basem Al-Shayeb. Al-Shayeb and research associate Rohan Sachdeva are co-first authors of the Nature paper.
New Cas protein
One of the huge phages also is able to make a protein analogous to the Cas9 protein that is part of the revolutionary tool CRISPR-Cas9 that Jennifer Doudna of UC Berkeley and her European colleague, Emmanuelle Charpentier, adapted for gene-editing. The team dubbed this tiny protein CasØ, because the Greek letter Ø, or phi, has traditionally been used to denote bacteriophage.
“In these huge phages, there is a lot of potential for finding new tools for genome engineering,” Sachdeva said. “A lot of the genes we found are unknown, they don’t have a putative function and may be a source of new proteins for industrial, medical or agricultural applications.”
Aside from providing new insight into the constant warfare between phages and bacteria, the new findings also have implications for human disease. Viruses, in general, carry genes between cells, including genes that confer resistance to antibiotics. And since phages occur wherever bacteria and Archaea live, including the human gut microbiome, they can carry damaging genes into the bacteria that colonize humans.
“Some diseases are caused indirectly by phages, because phages move around genes involved in pathogenesis and antibiotic resistance,” said Banfield, who is also director of microbial research at the Innovative Genomics Institute (IGI) and a CZ Biohub investigator. “And the larger the genome, the larger the capacity you have to move around those sorts of genes, and the higher the probability that you will be able to deliver undesirable genes to bacteria in human microbiomes.”
Sequencing Earth’s biomes
For more than 15 years, Banfield has been exploring the diversity of bacteria, Archaea — which, she says, are fascinating cousins of bacteria — and phages in different environments around the planet. She does this by sequencing all the DNA in a sample and then piecing the fragments together to assemble draft genomes or, in some cases, fully curated genomes of never-before-seen microbes.
In the process, she has found that many of the new microbes have extremely tiny genomes, seemingly insufficient to sustain independent life. Instead, they appear to depend on other bacteria and archaea to survive.
One year ago, she reported that some of the largest phages, a group she called Lak phages, can be found in our guts and mouths, where they prey on gut and saliva microbiomes.
The new Nature paper came out of a more thorough search for huge phages within all the metagenomic sequences Banfield has accumulated, plus new metagenomes provided by research collaborators around the globe. The metagenomes came from baboons, pigs, Alaskan moose, soil samples, oceans, rivers, lakes and groundwater, and included Bangladeshis who had been drinking arsenic-tainted water.
The team identified 351 phage genomes that were more than 200 kilobases long, four times the average phage genome length of 50 kilobytes (kb). They were able to establish the exact length of 175 phage genomes; the others could be much larger than 200 kb. One of the complete genomes, 735,000 base-pairs long, is now the largest known phage genome.
While most of the genes in these huge phages code for unknown proteins, the researchers were able to identify genes that code for proteins critical to the machinery, called the ribosome, that translates messenger RNA into protein. Such genes are not typically found in viruses, only in bacteria or archaea.
The researchers found many genes for transfer RNAs, which carry amino acids to the ribosome to be incorporated into new proteins; genes for proteins that load and regulate tRNAs; genes for proteins that turn on translation and even pieces of the ribosome itself.
“Typically, what separates life from non-life is to have ribosomes and the ability to do translation; that is one of the major defining features that separate viruses and bacteria, non-life and life,” Sachdeva said. “Some large phages have a lot of this translational machinery, so they are blurring the line a bit.”
Huge phages likely use these genes to redirect the ribosomes to make more copies of their own proteins at the expense of bacterial proteins. Some huge phages also have alternative genetic codes, the nucleic acid triplets that code for a specific amino acid, which could confuse the bacterial ribosome that decodes RNA.
In addition, some of the newly discovered huge phages carry genes for variants of the Cas proteins found in a variety of bacterial CRISPR systems, such as the Cas9, Cas12, CasX and CasY families. CasØ is a variant of the Cas12 family. Some of the huge phages also have CRISPR arrays, which are areas of the bacterial genome where snippets of viral DNA are stored for future reference, allowing bacteria to recognize returning phages and to mobilize their Cas proteins to target and cut them up.
“The high-level conclusion is that phages with large genomes are quite prominent across Earth’s ecosystems, they are not a peculiarity of one ecosystem,” Banfield said. “And phages which have large genomes are related, which means that these are established lineages with a long history of large genome size. Having large genomes is one successful strategy for existence, and a strategy we know very little about.”
The researchers divided the 351 megaphages into 10 new groups, or clades, named after words for “big” in the languages of the paper’s co-authors: Mahaphage (Sanskrit), Kabirphage, Dakhmphage and Jabbarphage (Arabic); Kyodaiphage (Japanese); Biggiephage (Australian), Whopperphage (American); Judaphage (Chinese), Enormephage (French); and Kaempephage (Danish).
The UC Berkeley work was primarily supported by Innovative Genomics Institute (IGI) and the National Institutes of Health. Of the 45 co-authors, 35 contributed to the research while affiliated with UC Berkeley: Banfield, Al-Shayeb, Sachdeva, Lin-Xing Chen, Fred Ward, Audra Devoto, Cindy Castelle, Matthew Olm, Keith Bouma-Gregson, Christine He, Raphaël Méheust, Brandon Brooks, Alex Thomas, Adi Lavy, Paula Matheus-Carnevali, Jennifer Doudna, Allison Sharrar, Alexander Jaffe, Rose Kantor, Ray Keren, Katherine Lane, Ibrahim Farag, Shufei Lei, Kari Finstad, Ronald Amundson, Karthik Anantharaman, Alexander Probst, Mary Power and Jamie Cate.