[ad_1]
When Chlamydia trachomatis, the bacterium that causes one of the most common sexually transmitted infections worldwide, infects a human cell, it hijacks parts of the host to build protective layers around itself.
Inside this makeshift fortress, the bug grows and reproduces, eventually bursting out in search of a new target and killing the host cell. While scientists have known for years that Chlamydia protects itself in this way, they were missing the mechanics until now.
Researchers from Duke University and the MRC Laboratory of Molecular Biology in Cambridge, UK, have shown that one Chlamydia protein, known as ChlaDUB1, is capable of manipulating human cells in two different ways, at least one of which appears to be essential for thriving inside its host.
The findings which appeared this week in Nature Microbiology could pave the way for treating Chlamydia with fewer antibiotics.
Structural biologists led by David Komander of the MRC Laboratory of Molecular Biology and Chlamydia experts at Duke University collaborated on the study. Initially, Komander and postdoctoral fellow Jonathan Pruneda, now an assistant professor at Oregon Health & Science University, contacted Duke Professor Raphael Valdivia, Vice Dean for Basic Science, to discuss the ChlaDUB1 protein, which Valdivia’s team had worked on before.
ChlaDUB1 is one of a class of proteins generated by Chlamydia to disrupt host cell function. Komander, Pruneda, and colleagues found that the protein is an enzyme, a deubiquitinase, that removes ubiquitin, a small protein that human cells attach to other proteins to activate them or to indicate that those proteins should be torn up. Human cells use ubiquitin to send signals, many of which are important for inflammatory responses to pathogens like Chlamydia.
Komander’s group determined through further study of the ChlaDUB1 enzyme’s shape that it can also modify proteins with acetylation to disrupt the alarms human cells raise to fight infection.
“Instead of making two proteins, one that has the deubiquitinase activity and a separate one that has acetylation activity, they’ve combined that into the same protein,” said coauthor Robert Bastidas, a research assistant professor who is part of Valdivia’s group at Duke.
Chlamydia is unlike other bacteria in that it can’t survive on its own outside of a human cell, Bastidas explained. He said it is likely that the bug has cast off large parts of its genome in order to better survive inside host cells. He hypothesizes that the bacterium saves space with this mashed-together protein, the only Chlamydia protein that has been found to have these two functions.
While it was clear that ChlaDUB1 was capable of both functions, Bastidas and his colleagues at Duke wanted to know what the enzyme was doing inside its host during Chlamydia infection. The researchers infected human cells with wildtype Chlamydia, as well as with mutant strains harboring defective copies of ChlaDUB1.
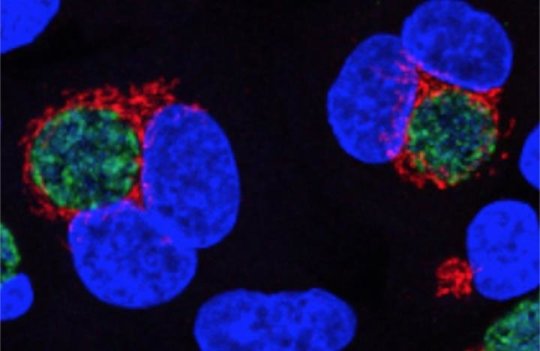
Once Chlamydia has built its fortress within the host cell, it breaks up the host cell’s Golgi apparatus and maneuvers the pieces around itself. The Golgi apparatus is a cellular compartment that typically stays close to the nucleus of the cell and modifies proteins by adding sugars that serve as baggage tags indicating whether the proteins should go to the plasma membrane or to some other cellular compartment. It’s not clear why the bacterium surrounds itself with pieces of the Golgi, perhaps to use the sugars and fats for its own growth, but it’s the only bacterium known to do so.
In the scientists’ infection trials, the wildtype Chlamydia chopped up the Golgi as usual. But when infected with a bug carrying a mutant enzyme, the human cells’ Golgi remained intact, suggesting that ChlaDUB1’s activity is necessary for this aspect of Chlamydia infection.
Bastidas also hypothesizes that the ability for ChlaDUB1 to remove ubiquitin from host proteins protects Chlamydia from the host inflammatory response.
Next, the researchers want to find a drug that will specifically disrupt the function of ChlaDUB1, thus slowing the bacteria’s ability to fight off attack by the host immune system. “If we develop these inhibitors and they’re specific enough, then we won’t have to use antibiotics” or at least use fewer of them, said Bastidas.
In a world where antibiotic use can lead to antibiotic resistance or to disruption of the delicate microbiome of the vagina and the urinary tract, where Chlamydia prefers to reside, Bastidas says a more tailored therapy could prove a better tool to fight infection.
This research was supported by the EU FP7 infrastructure grant BIOSTRUCT-X (283570), the UK Medical Research Council (U105192732), the European Research Council (724804), the Lister Institute for Preventive Medicine, an EMBO Long-Term Fellowship, the National Institutes of Health (R01AI100759), the National Institute of Allergy and Infectious Diseases (STI CRC U19 AI084044), and North West Cancer Research.
[ad_2]