[ad_1]
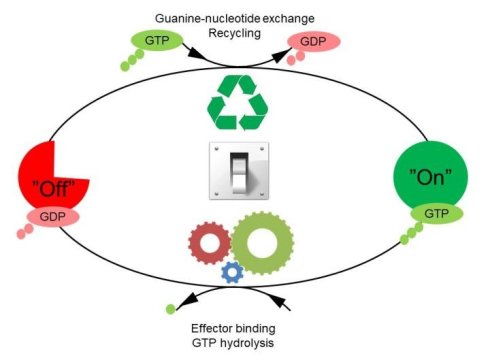
The GTPases constitute a very large protein family, whose members are involved in the control of cell growth, transport of molecules, synthesis of other proteins, etc. Despite the many functions of the GTPases, they follow a common cyclic pattern. The activity of the GTPases is regulated by factors that control the ability of GTPases to bind and hydrolyse guanosine triphosphate (GTP) to guanosine diphosphate (GDP). So far, it has been the general assumption that a GTPase is active or “on” when it is bound to GTP and inactive or “off” in complex with GDP. The GTPases are therefore sometimes referred to as molecular “switches.”
The bacterial translational elongation factor EF-Tu is a GTPase, which plays a crucial role during the synthesis of proteins in bacteria, as the factor transports the amino acids that build up a cell’s proteins to the cellular protein synthesis factory, the ribosome. Previous structural studies using X-ray crystallography have shown that EF-Tu occurs in two markedly different three-dimensional shapes depending on whether the factor is “on” (i.e. bound to GTP) or “off” (i.e. bound to GDP). The binding of GTP/GDP have therefore always been thought to be decisive for the factor’s structural conformation.
However, a research collaboration between researchers from the Department of Molecular Biology and Genetics at Aarhus University and two American universities reveals that EF-Tu’s — and probably also other GTPases — structure and function are far more complex than previously assumed. In Søren Thirup’s group, X-ray crystallographic analysis of E. coli EF-Tu has shown that EF-Tu bound to a variant of GTP, GDPNP, can also occur in the “off” state, which is characterised by a more open structure. In collaboration with American researchers, Charlotte Knudsen’s PhD student, Darius Kavaliauskas, conducted further studies using a special form of fluorescence microscopy that makes it possible to observe the spatial structure of individual EF-Tu molecules in solution.
EF-Tu was labelled with a fluorescence donor and a fluorescence acceptor. When the donor is irradiated with light of a certain wavelength, the light will be absorbed and converted to light with a new wavelength. The acceptor will capture the light and reemit it at a third wavelength, if it is in close proximity to the donor. The transmitted light is measured in a confocal microscope, whereby the distance between donor and acceptor in the EF-Tu molecules can be determined for thousands of molecules in solution, thereby providing information about the dynamic aspects of EF-Tu.
GTPases are far more dynamic than assumed
The study showed that EF-Tu in solution is not in a fixed structure, when the factor is bound to GDP or variations with GDPNP, and thus should be “off” or “on,” respectively. Instead, EF-Tu turned out to be extremely dynamic by appearing as a mixture of structures. This tendency was most pronounced when GDPNP was included in the solution, in accordance with the X-ray crystallographic study. Only when binding to the ribosome, EF-Tu assumed the expected active form.
The results indicate that in the future, GTPases should be regarded as much more flexible molecules that are not only “on” or “off.” GTPases are obvious drug targets: as examples, bacterial infections can in principle be cured by inhibition of EF-Tu, while the GTPase ras p21 is misregulated in approximately 30% of all cancers — especially the particularly fatal forms in the lung, colon and pancreas. However, so far it has not been possible to develop a usable drug against these two targets, but the discovery of the high flexibility of the GTPases may help to change this.
Story Source:
Materials provided by Aarhus University. Note: Content may be edited for style and length.
[ad_2]