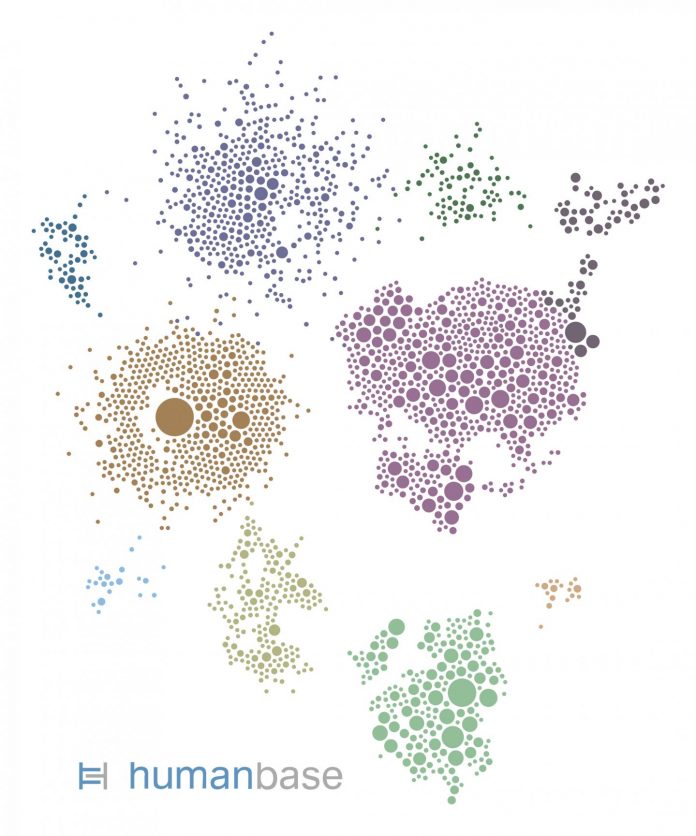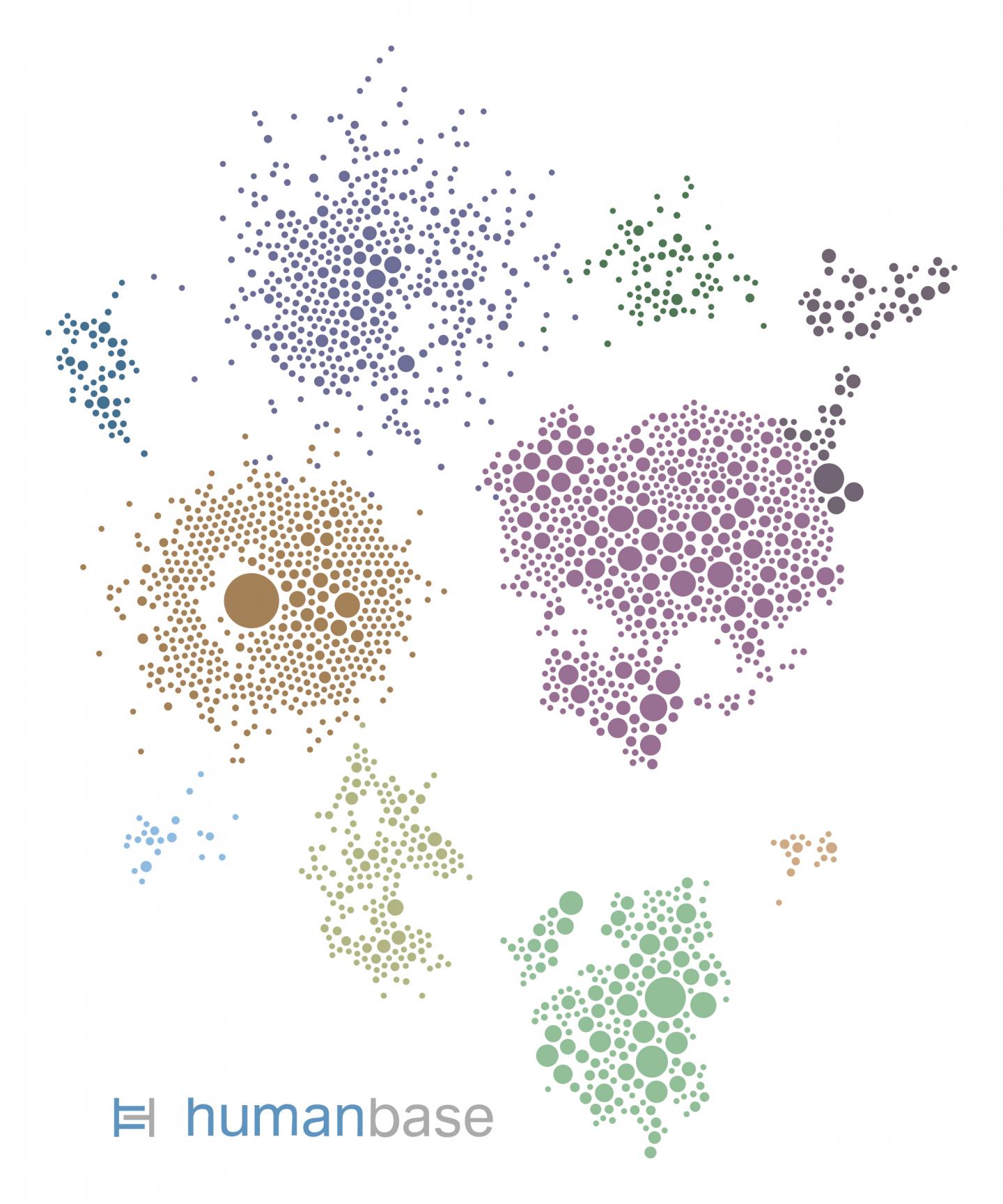
Photo: This gene network was generated by HumanBase for the kidney cell type most vulnerable to SARS-CoV-2 infection. Each circle represents a gene expressed by the cells. Each gene is color-coded…
view more
Credit Image: hb.flatironinstitute.org/covid-kidney
People with diabetes — especially the 20 to 40 percent with diabetic kidney disease — are among the most at risk for serious complications and death from COVID-19. A new study of gene expression utilizing machine learning peered inside the kidney cells of COVID-19 patients and diabetic kidney disease patients and made a surprising discovery: Similar molecular processes were activated in both sets of patients, revealing potential avenues of viral vulnerability. The researchers report their findings in Kidney International.
The results suggest that diabetes may predispose patients to infection with the novel coronavirus or to more severe COVID-19 by spurring biological processes used by the virus to infect and replicate. The same study found that medications commonly used to treat hypertension and diabetic kidney disease probably don’t increase coronavirus infection risk, despite initial concerns about drug interactions with viral entry pathways.
“Our hypothesis is that diabetes primes these kidney cells in some way that makes them especially vulnerable to COVID,” says study co-author Aaron Wong, a data scientist and project leader at the Flatiron Institute’s Center for Computational Biology (CCB) in New York City. The new findings will help guide strategies to reduce the risk of COVID-19 to diabetics, he says.
The research was led by Olga Troyanskaya, deputy director of genomics at the CCB and a professor at Princeton University, and Matthias Kretzler, a nephrologist and a professor at the University of Michigan’s academic medical center, Michigan Medicine. Wong is co-lead author of the study along with the CCB’s Rachel Sealfon, Princeton’s Chandra Theesfeld, and Michigan Medicine’s Rajasree Menon, Edgar Otto and Viji Nair.
The coronavirus responsible for COVID-19, dubbed SARS-CoV-2, is best known for targeting cells in the respiratory system. However, the virus can also invade cells in organs such as the heart and kidneys. The virus infiltrates cells by latching on to a protein called angiotensin-converting enzyme 2, or ACE2. This viral piggybacking prompted the hypothesis that cells that produce lots of ACE2 are more vulnerable to infection. The researchers set out to find which kidney cells had elevated ACE2 levels and what made those cells extra vulnerable in diabetic kidney disease patients.
The researchers started by measuring ACE2 levels and gene expression in cells taken from biopsies from healthy kidney donors and patients with diabetic kidney disease. Kidney biopsies weren’t an option for COVID-19 patients, so the researchers looked instead at kidney cells in urine samples from COVID-19 patients with kidney damage. These provided a unique look inside the organ without invasive procedures. Analyzing the samples, the researchers found ACE2 expression primarily in proximal tubule epithelial cells. These cells play a major role in the reabsorption of substances such as water, salt, glucose and amino acids by the kidneys.
Only around one-fifth of the proximal tubule epithelial cells had detectable ACE2 and were therefore vulnerable to infection. So what made those cells so vulnerable to COVID-19 in diabetic kidney disease patients? To find out, the researchers looked at which biological processes ramp up in ACE2-expressing cells. To identify these pathways in the vast complexity of cellular circuitry, researchers used the HumanBase.io system developed by Troyanskaya’s group at the CCB. HumanBase uses machine learning to identify biological connections between genes and pinpoint functionally related groups of genes (called modules) in the context of specific tissues, cell types and diseases. “HumanBase is what made this analysis possible,” Troyanskaya says.
HumanBase identified several modules linked to ACE2 production in COVID-19 patients. Those modules were associated with processes related to a cell’s reaction to viral invasion, the activation of immune responses, and viral replication. Surprisingly, those same modules were also present in patients with diabetic kidney disease. “The cells in the diabetic kidney patients looked a lot like the cells in the COVID-19 patients,” says Sealfon, “with many genes upregulated that respond to or are engaged by viruses.”
The consequences are twofold: Diabetic kidney disease could make cells more vulnerable to the coronavirus, and diabetic kidney disease and COVID-19 may compound each other’s effects, potentially triggering a severe immune response that causes kidney damage.
The researchers also found that ACE2 levels were unaffected by drugs known as renin-angiotensin-aldosterone system inhibitors, which are commonly used to treat hypertension and diabetic kidney disease. These drugs modulate ACE or inhibit the pathway used by the protein. “At the beginning of the pandemic, there was a back-and-forth about whether patients should stop taking these ACE pathway modulators because they might increase the risk of infection,” Theesfeld says. “Now a few studies, including ours, show that they should keep taking their medicines.”
The results come with a major caveat, though. The study looked at the gene expression in kidney cells from patients with diabetes or COVID-19, but not both. More work is needed to confirm how the two illnesses behave in tandem. The researchers say that the next step is to look at kidney tissues affected by both diabetic kidney disease and COVID-19. Such a study could help identify whether specific therapies could reduce COVID-19 susceptibility and progression in diabetic kidney disease patients.
###
ABOUT THE FLATIRON INSTITUTE
The Flatiron Institute is the research division of the Simons Foundation. The institute’s mission is to advance scientific research through computational methods, including data analysis, modeling and simulation. The institute’s Center for Computational Biology develops new and innovative methods of examining data in the biological sciences whose scale and complexity have historically resisted analysis. The center’s mission is to develop modeling tools and theory for understanding biological processes and to create computational frameworks that will enable the analysis of the large, complex data sets being generated by new experimental technologies.
TDnews (tunisiesoir.com)