[ad_1]
Proteins — the molecular machines that drive the processes underlying biology — are made from just 20 canonical building blocks called amino acids. For nearly two decades, scientists have sought methods to engineer new amino acids to build proteins.
A team of Boston College chemists have developed a technology to precisely incorporate a range of useful non-canonical amino acids into proteins made in eukaryotes, the broad class of higher organisms that includes humans, the team reported in the journal Cell Chemical Biology.
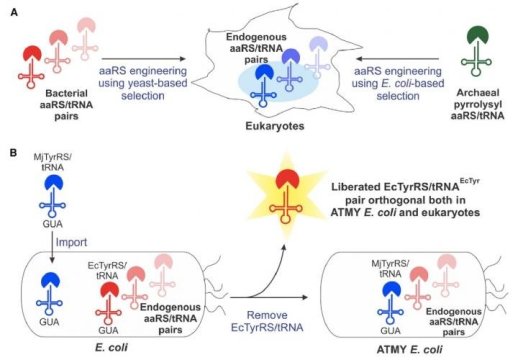
Approximately 15 years ago, scientists first saw the potential of an engineered, bacteria-derived genetic machinery — known as an aminoacyl-tRNA synthetase/tRNA pair — for incorporating non-canonical amino acids into proteins produced in eukaryotic cells. But the method has faced a number of technical restrictions that limited its widespread development.
The Boston College team overcame many of those limitations by developing a novel strain of E. coli bacteria that enables facile engineering of the bacteria-derived aminoacyl-tRNA synthetase/tRNA pair, according to Assistant Professor of Chemistry Abhishek Chatterjee, who led the project. This new approach enabled the incorporation of various non-canonical amino acids, including p-boronophenylalanine, into proteins produced in human cells as well as in the engineered strain of E. coli.
Chatterjee said the team was surprised by the facility of the new approach, which is described in the new report “Resurrecting the bacterial tyrosyl-tRNA synthetase/tRNA pair for expanding the genetic code of both E. coli and eukaryotes.”
“Creating this novel E. coli strain required substituting its native aminoacyl-tRNA synthetase/tRNA pair with a counterpart from a different organism, which we anticipated would be very difficult,” he said. “But it turned out to be quite feasible. That opens up this complete technology.”
Chatterjee said the team sought to create a new method to engineer and monitor protein functions as a way of expanding the scientific understanding of the processes that guide protein functions in our cells.
“Thousands of proteins are encoded in the genome that make us who we are, but we know very little about that process,” said Chatterjee. “In human cells, there are roughly 20,000 protein-coding genes. What they are doing and how they are doing it remains difficult to study. One of the major problems is that if you want to know what they are doing, you have to spy on them. You need to attach a probe that can report back on what is going on.”
Introducing such probes has proven difficult, as the process often damages the target protein.
In any cell, proteins are made from 20 amino acids — a fixed group guided into order by genetic instructions.
“The idea is that we can introduce a new building block into proteins that nature does not have — beyond the 20 canonical amino acids that nature uses,” Chatterjee said. “If we can do that, we have the ability to very specifically introduce a wide variety of non-natural functionalities into any site of virtually any protein.”
The immediate benefit would be to assist researchers who are still unraveling the mysteries of cell biology and protein function.
“You could create a protein with a non-canonical amino acid into any chosen site, load it with probes that are very tiny and give out an optical signal that tells where it is going,” Chatterjee said. “It could allow you to manipulate how the protein is working. You could introduce limits, so whatever the protein is doing, it can’t do any more. And you could remove the probe by using an external signal such as light. This technology opens up numerous new ways one can start to probe and engineer protein function, which would be very challenging otherwise.”
Story Source:
Materials provided by Boston College. Note: Content may be edited for style and length.
[ad_2]