[ad_1]
When bacteria enter the digestive tracts of their hosts, including humans, they encounter a highly acidic environment. Bacteria have evolved elegant mechanisms to survive and colonize this habitat, such as highly resistant and impermeable outer membranes and systems to pump out any toxins that make it beyond that barrier. However, the molecular mechanisms behind many of these processes have remained unclear.
As part of an international effort, a research team centered at Osaka University has revealed how Salmonella bacteria use a protein called RamR to sense the presence of bile acids in their surroundings. This induces a cascade of events that help the bacteria to adapt to the difficult conditions in the gut.
In this work, reported in the journal Scientific Reports, the team first analyzed a regular strain of Salmonella and one in which RamR had been deleted. As it was already known that RamR represses the expression of the gene RamA, which is linked to the export of substances from bacterial cells, they compared RamA expression in the two sets of bacteria upon exposure to different types of bile acid. The results showed that two particular bile acids common in the gut caused dramatic increases of RamA expression in the regular Salmonella, but not in the RamR mutant.
“We already knew that Salmonella starts to express many genes when reaching the gut, but these findings clearly showed that RamR is a key part of its adaptation,” corresponding author Kunihiko Nishino says. “We then used surface plasmon resonance analysis to show that RamR binds directly to the primary bile acids cholic acid and chenodeoxycholic acid.”
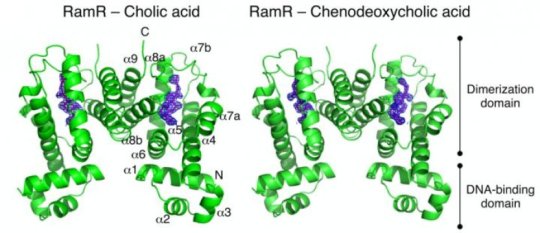
To obtain more details about this binding and its downstream effects, the team then determined the crystal structures of RamR when bound to each of these acids. In these complexes, they identified the formation of hydrogen bonds and the associated uncoiling of an alpha-helix domain in RamR, both of which are novel binding mechanisms for this protein.
“When RamR is bound in this way, it loses some DNA binding affinity and can no longer repress its RamA target,” lead author Suguru Yamasaki says. “This leaves the RamA protein free to activate the AcrAB-TolC efflux system, which pumps damaging substances out of bacterial cells and helps them to survive in the small intestine and gallbladder.”
These findings about RamR suggest that it could be a good target for developing new treatments against infections of gut bacteria or for rebalancing the bacterial population in the gut to ensure better intestinal function.
Story Source:
Materials provided by Osaka University. Note: Content may be edited for style and length.
[ad_2]