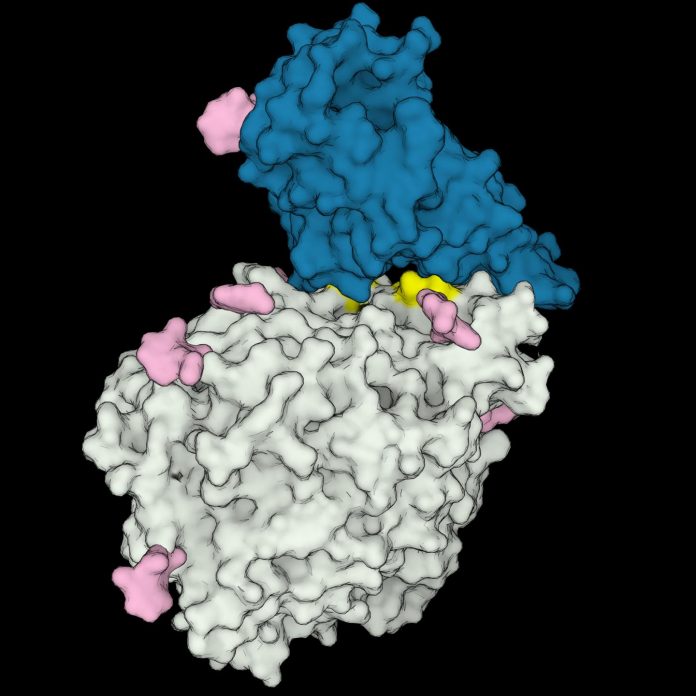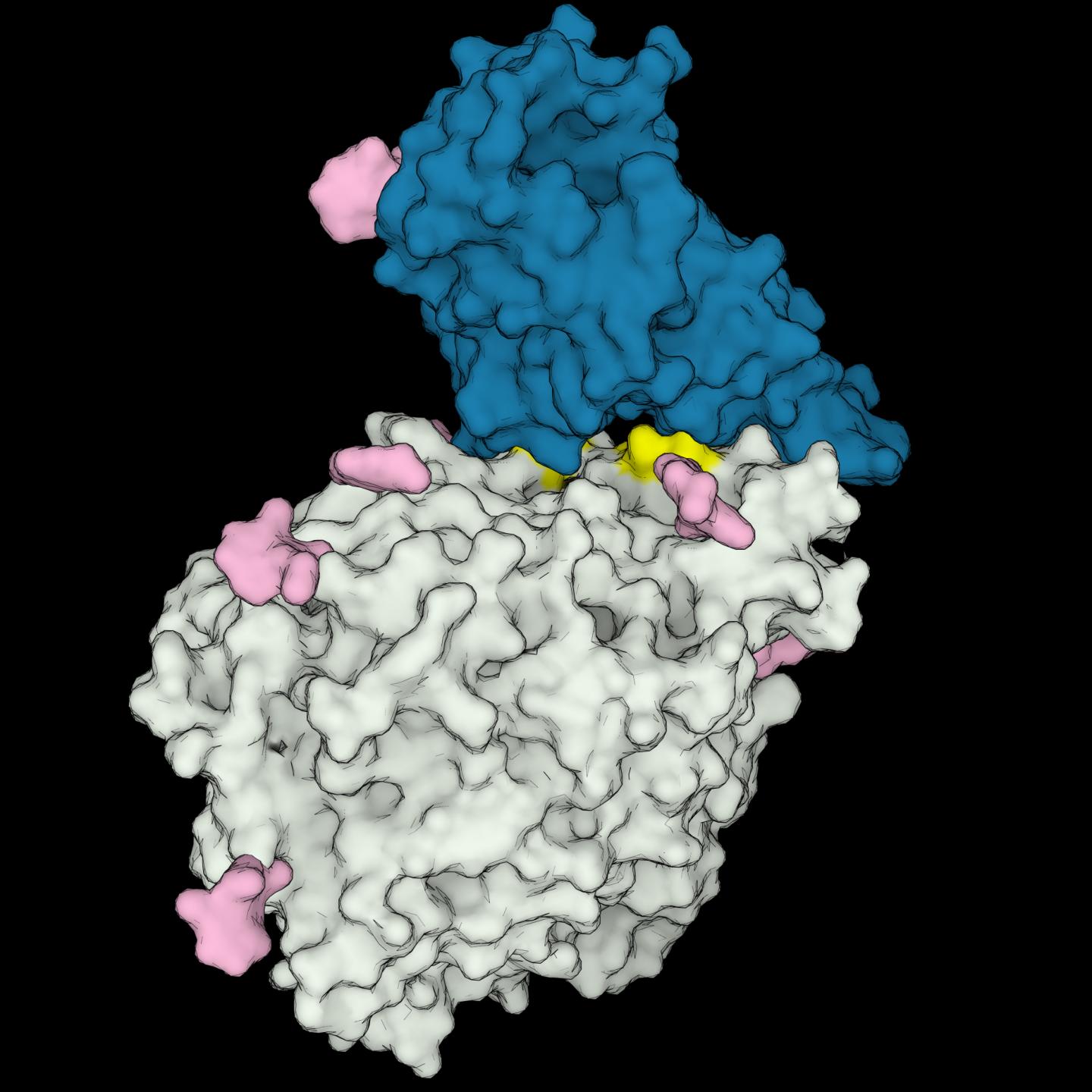
Photo: 3D structure model of the receptor-binding domain of SARS-CoV-2 (in blue) interacting with the human ACE2 receptor (in gray). Amino acids important to the interaction, which are present only in…
view more
Credit Image: Rodrigues et al. 2020 (CC-BY 2.0)
Some animals are more susceptible to Covid-19 infection than others, and new research suggests this may be due to distinctive structural features of a protein found on the surface of animal cells. João Rodrigues of Stanford University, California, and colleagues present these findings in the open-access journal PLOS Computational Biology.
Previous research suggests that the current pandemic began when the virus that causes Covid-19, SARS-CoV-2, jumped from bats or pangolins to humans. Certain other animals, such as cattle and cats, appear to be susceptible to Covid-19, while others, such as pigs and chickens, are not. One zoo even reported infections in tigers. However, it was unclear why some animals are immune and others are not.
To address this question, Rodrigues and colleagues looked for clues in the first step of infection, when SARS-CoV-2’s “spike” protein binds to an “ACE2” receptor protein on the surface of an animal cell. They used computers to simulate the proteins’ 3D structures and investigate how the spike protein interacts with different animals’ ACE2 receptors–similar to checking which locks fit a certain key.
The researchers found that certain animals’ ACE2 “locks” fit the viral “key” better, and that these animals, including humans, are susceptible to infection. Despite being approximations, the simulations pinpointed certain structural features unique to the ACE2 receptors of these susceptible species. The analysis suggest that other species are immune because their ACE2 receptors lack these features, leading to weaker interactions with spike proteins.
These findings could aid development of antiviral strategies that use artificial “locks” to trap the virus and prevent it from interacting with human receptors. They could also help improve models to monitor animal hosts from which a virus could potentially jump to humans, ultimately preventing future outbreaks.
“Thanks to open-access data, preprints, and freely available academic software, we went from wondering if tigers could catch Covid-19 to having 3D models of protein structures offering a possible explanation as to why that is the case in just a few weeks,” Rodrigues says.
His team plans to continue refining the computational tools used in this study.
###
Research Article
Peer-reviewed; Simulation / modeling; N/A
In your coverage please use this URL to provide access to the freely available article in PLOS Computational Biology:
https:/
Citation: Rodrigues JPGLM, Barrera-Vilarmau S, M. C. Teixeira J, Sorokina M, Seckel E, Kastritis PL, et al. (2020) Insights on cross-species transmission of SARS-CoV-2 from structural modeling. PLoS Comput Biol 16(12): e1008449. https:/
Author Affiliations:
Department of Structural Biology, Stanford University School of Medicine, Stanford, California, United States of America
Institute of Advanced Chemistry of Catalonia (IQAC), CSIC, Barcelona, Spain
Program in Molecular Medicine, Hospital for Sick Children, Toronto, Ontario, Canada
ZIK HALOMEM & Institute of Biochemistry and Biotechnology, Martin Luther University Halle-Wittenberg, Biozentrum, Halle (Saale), Germany
Department of Obstetrics and Gynecology, Stanford University School of Medicine, Stanford, California, United States of America
Image Caption: 3D structure model of the receptor-binding domain of SARS-CoV-2 (in blue) interacting with the human ACE2 receptor (in gray). Amino acids important to the interaction, which are present only in COVID-susceptible animal species are highlighted in yellow. Sugars bound to the proteins are shown in pink.
Image Credit Image: Rodrigues et al. 2020 (CC-BY 2.0)
Image Link: https:/
Funding: JPGLMR acknowledges support from the Molecular Sciences Software Institute (ACI-1547580) (https:/
Competing Interests: The authors have declared that no competing interests exist.
TDnews (tunisiesoir.com)