[ad_1]
The proteins in our bodies are sophisticated structures that perform specific jobs to keep us functioning and healthy. In many cases, these tiny machines are switched on or off through a two-step process where one part of the protein sends messages to another part called the “active site,” triggering the protein to start or stop its job. Sometimes this process — known as allostery — is disrupted, which leads to or contributes to the development of certain diseases.
Understanding how a protein is wired could help researchers develop ways to control its activity, and scientists at the Advanced Science Research Center (ASRC) at The Graduate Center, CUNY, believe they’ve come up with a reliable way to determine this, according to a newly published study in eLife.
“Just like it’s hard to guess how a light switch is wired to a light bulb in a room without seeing behind the walls, it’s hard to predict what remote area of a protein are wired to its active site without seeing the details inside the structure,” said Daniel Keedy, assistant professor with the ASRC’s Structural Biology Initiative and with The City College of New York’s chemistry and biochemistry departments.
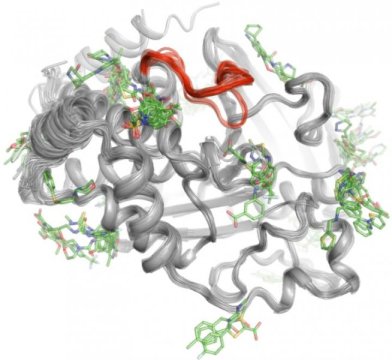
To solve this mystery, Keedy and his colleagues explored the signaling capabilities of protein tyrosine phosphatase 1B (PTP1B), which researchers believe plays a significant role in type 2 diabetes and possibly breast cancer. They first used X-ray crystallography at different temperatures to see how atoms inside the protein move. This revealed specific areas where the atoms move like windshield wipers or a Newton’s cradle toy to send messages. Next, the team performed high-throughput experiments with the protein to determine which small molecules bind to these signaling sites.
“This two-step process allowed us to see not only where the signaling originates inside PTP1B, but also what small molecules are capable of sending messages to the active site. This knowledge could one day help us develop therapies that send specific messages to control a protein’s activity and disrupt the development of type 2 diabetes,” said Keedy.
“I am quite keen on the potential applications of Daniel’s work,” said Kevin Gardner, director of the ASRC’s Structural Biology Initiative and Einstein Professor of Chemistry and Biochemistry at City College. “This technique provides a powerful experimental route to asking where small molecules can bind to protein targets, enabling drug discovery and development by showing where one might be able to find previously unanticipated ‘footholds’ for compounds to bind and control protein activity.”
Keedy and his colleagues have so far only looked at PTP1B, but their method uses readily available technologies that can be used to study the process of allostery in many other important proteins within our bodies. They hope to gain a new understanding of how the functions of these other proteins are controlled, which could be useful for future development of new drug therapies.
Story Source:
Materials provided by Advanced Science Research Center, GC/CUNY. Note: Content may be edited for style and length.
[ad_2]