[ad_1]
A powerful new tool is available for neuroscience investigation into brain development, the mechanisms of memory and learning, and the role of brain dysregulation in neuropsychiatric diseases like addiction, depression, schizophrenia and Alzheimer’s disease.
This molecular biology tool — developed and validated by University of Alabama at Birmingham researchers — is able to selectively and robustly turn on genes in neurons. It has been delivered to neurons growing in a dish, and more importantly, it has also been delivered to brain neurons in living adult rats. The tool can increase the expression of a single gene or the expression of multiple genes at the same time, and the amount of increased gene expression can be controlled.
This selective ability to increase gene expression — which means the gene makes more mRNA that codes for more protein — enables researchers to query the role of individual genes or groups of genes.
Such queries are building blocks to learn how expression of different gene programs during development controls brain growth, wiring and the types of neurons created. For the adult brain, the tool can help explain how gene programs altered by neural activity and behavioral experience lead to adaptive behavior. The tool can also help investigate dysregulation of gene programs — many of which are implicated in neuropsychiatric diseases — that occur during brain development or in the fully developed adult brain.
The tool uses well-known CRISPR-based technology, which was packaged into lentiviruses for stable introduction into neurons.
“CRISPR-based technology has provided new avenues to interrogate gene function, but difficulties in transgene expression in post-mitotic neurons have delayed incorporation of these tools in the central nervous system,” said Jeremy Day, Ph.D., leader of the UAB research team. “Here, we demonstrate a highly efficient, neuron-optimized dual lentiviral CRISPR-based transcriptional activation system capable of robust, modular and tunable gene induction and multiplexed gene regulation across several primary rodent neuron culture systems.”
Day is an assistant professor in the Department of Neurobiology, UAB School of Medicine, and a scientist in the UAB Civitan International Research Center. The study is published in the journal eNeuro, with Katherine E. Savell and Svitlana V. Bach, Ph.D., UAB Department of Neurobiology, as co-first authors.
“With the development of CRISPR technology, we became able to selectively edit genes and gene expression levels on demand,” said Erin Calipari, Ph.D., an assistant professor in the Vanderbilt Brain Institute at Vanderbilt University, who was not involved in the study. “This provided a powerful new tool for linking the effects of gene expression and gene regulation to cellular function and behavior; but until now, these systems were primarily optimized for use in cell culture.”
“While it sounds trivial,” Calipari said, “taking a genetic tool from culture to in vivo use is incredibly difficult. This technology is going to revolutionize the use of these systems in vivo in order to link changes in transcriptional regulators at specific genes in defined brain regions to neural activity and behavior. Furthermore, these tools are going to allow for the region-specific manipulation of individual genes, or groups of genes, in awake and behaving animals, which is going to give us an unprecedented view of the role of gene expression in behavior.”
“Understanding the basic mechanisms governing this process,” she said, “is the first step to developing selective treatments for psychiatric disease.”
Study details
The CRISPR/Cas9 system is widely known as molecular scissors that can cut at a precise site in the genome to remove a gene or add a gene. An inactive form of CRISPR/Cas9 that is unable to cut DNA can be joined with transcriptional effectors to activate expression of a gene at a precise location. Day and colleagues used this system, called CRISPRa. They packaged it into lentiviruses to allow stable expression of CRISPR machinery, allowing researchers to test its ability to regulate genes in neurons. The system includes a CRISPR-defective-Cas9 with activators and a single guide RNA that guides the CRISPR-dCas9 to a precise point on the genome.
One limitation of using CRISPRa in neurons is that promoters that are commonly used to drive molecular machinery in other types of cells were ineffective at driving CRISPR expression in neurons. To circumvent this, the UAB researchers selected a neuron-specific promoter, based on the promoter sequence for the human Synapsin, or SYN, gene, to express their CRISPRa tool in neurons. The UAB researchers then developed CRISPR single guide RNAs to target this system to a range of distinct genes, from small transcription factors to large extracellular proteins. They showed that this system significantly induced expression for each of these gene targets in primary rat neurons from the cortex, hippocampus and striatum of the brain.
Use of multiple single guide RNAs, called multiplexing, allowed tunable upregulation of single genes, or coordinated upregulation of multiple genes.
The UAB teams also tested the system on a very complex brain gene, the brain-derived neurotrophic factor, or Bdnf. Bdnf is extremely difficult to study due to its complex transcriptional regulation, yet it is important for its central role in diverse processes such as neuronal differentiation and survival, dendritic growth, synaptic development, long-term potentiation, and memory formation. Bdnf has nine different promoter locations, and each leads to a variant mRNA transcript.
“Due to this complexity, attempts to characterize distinct functional roles of individual Bdnf mRNAs in neurons have produced conflicting results, and currently available tools either lack the ability to selectively upregulate single Bdnf transcript variants or require cumbersome molecular cloning protocols to generate gene-specific targeting constructs,” Day said.
Day and colleagues were able to use their CRISPR-dCas9 system to specifically target single promoters at two different sites on Bndf. Each induced only the transcript for that site, without altering expression at any of the other promoters. They then characterized differentially expressed genes induced downstream by each unique transcript and measured accompanying changes in neuronal physiology.
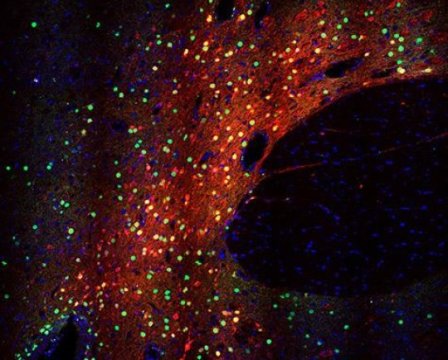
Finally, the researchers validated their gene regulation system in adult rats, using stereotactic surgery to infuse CRISPRa-carrying lentiviruses into the prefrontal cortex, hippocampus and nucleus accumbens of the brain.
“I think we are just beginning to scratch the surface of the types of gene regulation now possible through generation of effector-coupled dCas9 tools like these,” said Catherine Jensen Peña, Ph.D., an assistant professor at the Princeton Neuroscience Institute, who was not involved in the study. “This opens exciting possibilities in neuroscience, particularly for studying the complexity of brain function and brain disorders.”
Support came from National Institutes of Health grants DA039650, DA034681, MH114990, DA042514, MH112304 and DA041778; the Civitan International Research Center at UAB; and the UAB Pittman Scholars Program.
[ad_2]