[ad_1]
Yale scientists have taken a novel approach to unraveling the complex structure and regulation of enzymes: They Googled it.
In a new study published online this week in the Proceedings of the National Academy of Sciences, chemistry professor Victor Batista and his colleagues used the Google algorithm PageRank to identify key amino acids in the regulation of a bacterial enzyme essential for most microorganisms.
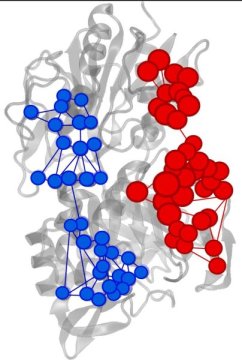
Enzymes are biomolecules with the unique capability of accelerating chemical reactions that are necessary for life. Although these chemical reactions normally take place in a small portion of the enzyme — known as the active site — the acceleration of the reaction is usually regulated by the binding of a molecule in a different part of the enzyme. The binding position is known as the allosteric site.
Despite decades of study, it is still poorly understood how information is transferred from the allosteric site to the active site. Much of the difficulty has to do with the large number of atoms involved and the great structural flexibility of enzymes.
The Yale team noted that a similar question had been addressed years earlier in the realm of computer science. Researchers at Google had studied the flow of information on the Internet, using PageRank to indicate the importance of each web page in terms of the number and quality of links to other Internet sites.
“This problem is completely analogous to the exchange of information between distant sites that characterizes allosterism,” said Uriel Morzan, a postdoctoral associate in Batista’s lab and co-first author of the study. “By finding out how the information flow through each atom changes with the binding of an allosteric activator to the enzyme, it is possible to find the information channels that are being activated.”
The Yale researchers identified important amino acids for the allosteric process in imidazole glycerol phosphate synthase (IGPS), a bacterial enzyme found in most microorganisms.
The research paves the way for additional experiments related to IGPS activity that may lead to the development of new antibiotics, pesticides, and herbicides.
“It’s exciting that data science methods are starting to percolate into the field of theoretical chemistry, providing new tools for understanding fundamental aspects of catalytic molecular systems when combined with state-of-the-art molecular dynamics simulations and nuclear magnetic resonance (NMR) spectroscopy,” said Batista, who is also a member of the Energy Sciences Institute at Yale’s West Campus.
Co-author J. Patrick Loria, a Yale professor of chemistry and of molecular biophysics and biochemistry, added: “It is the synergistic combination of experimental NMR and computational tools that enables this deeper insight into biological function and demonstrates the importance of collaboration between theorists and experimentalists.”
Story Source:
Materials provided by Yale University. Note: Content may be edited for style and length.
[ad_2]